Molecular epidemiology
The characterisation of microbial population diversity with sequencing is an active field of research in Microb’UP
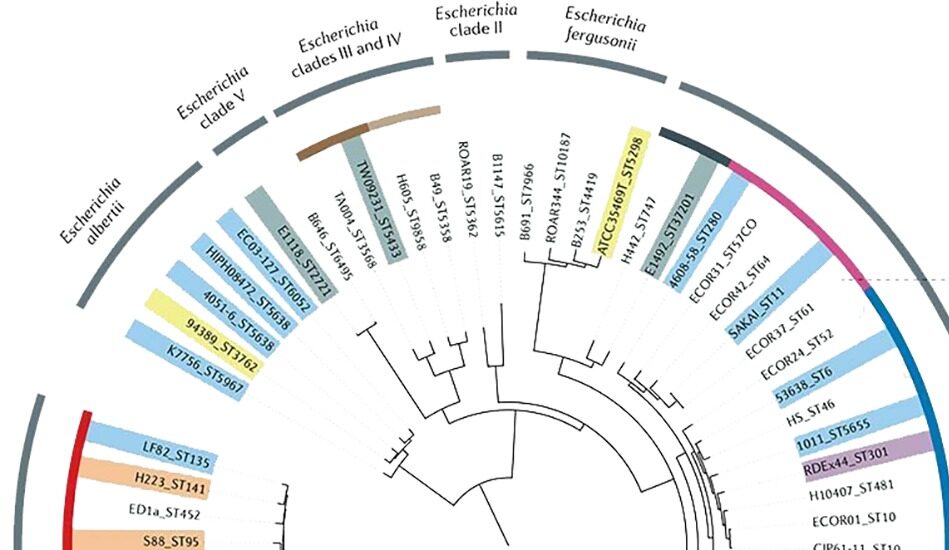
Phylogenetic tree of Escherichia coli from Denamur et al Nature Review Microbiology 2021
With the democratization of sequencing technologies, genomics has become a key element of the characterization of microbes and infectious diseases. Molecular epidemiology, with the study of a rising number of samples is offering snapshots of the diversity found within species. The benefit of full genome sequences is multiple. First, it allows fine scale analysis of the species phylogenies that are sometime not resolved for some microbes. Second, it allows to track the evolutionary dynamics occurring within species. At large scale, these dynamics are well illustrated with the propagation of a succession of SarsCov2 variant or with the spread of antibiotic resistance among bacterial species. But, full genome sequencing can also offer a high enough resolution to tackle viral or bacterial dynamics within a hospital or through a chain of contamination. Third, access to the full genome gives access to the functional elements of these microbes and can help identify genes or alleles associated with some clone successes, from antibiotic resistance or virulence genes, to patho-adaptative mutations.
With a vast number of teams with members directly involved in hospital virology, bacteriology and parasitology laboratory, Microb’UP network has access to a vast number of strains that are sequenced and studied in depth with the molecular epidemiology approach. On organisms ranging from HIV to Escherichia coli or sordariales. Modelers are also present in the Microb’UP to integrate these data and make inferences about the evolutionary processes at play.
News
MICROB’UP Webinar series : AI and Malaria Diagnosis
MICROB'UP Webinar Series: AI and Malaria Diagnosis This webinar has been recorded, find the link HERE Tuesday, February 17th 2026, at 11:00am (Paris) 11:00am: Gilles COTTRELL - MERIT, IRD, Inserm, Université Paris Cité: State of the art and presentation...

JeCCo 16ᵗʰ Annual Symposium: “How Science Adapts to a Changing Society”
The young researchers association of Institut Cochin (JeCCo) organizes its 16ᵗʰ Annual Symposium: “How Science Adapts to a Changing Society” on January 26th, 2026. The symposium will bring together researchers from diverse fields to discuss how science innovates...

MICROB’UP Webinar series : Spatial Organization and Functional Dynamics in Bacteria: From Cell Cycle to Host and Prey Interactions
MICROB'UP Webinar Series: Spatial Organization and Functional Dynamics in Bacteria: From Cell Cycle to Host and Prey Interactions This webinar has been recorded, find the link HERE Monday, January 12th 2026, at 11:00am (Paris) 11:00am: Christophe...
MICROB’UP Webinar series : Microbes and brain: intimate enemies, invisible allies
MICROB'UP Webinar Series: Microbes and Brain: Intimate Enemies, Invisible Allies This webinar has been recorded, find the link HERETuesday, December 9th, at 11:30am (Paris) 11:30am: Eliette BONNEFOY - Institut Cochin : Brain viral infection and Alzheimer's...
